Parameter reference
Description
This code takes in single cell intensity data from a deep dye drop microscopy experiment, determines the number of live and dead cells, and assigns each live cell to a phase of the cell cycle.
Input
Deep Dye Drop requires two inputs:
-
The name or path to the input object level data. The input
datacan either be a file (.txt, .csv, .tsv) or a folder containing one .txt file for each well. -
A python dict
ndictthat maps column names in the input data into a format compatible with the script.
Formatting
The input file should contain single cell data (each cell occupies a row) in a .cvs or .txt file. Plate (barcode), well, agent, concentration, timepoint, and role (negative control, positive control or treatment) identifiers as well as intensity measures for each channel should be in columns.
Output
By default, the cell cycle gating script will output a .pdf and .csv file for each plate analyzed. The .pdf file will contain EdU vs DNA content plots (one plot per well, each point shows a single cell) with the cell cycle gates overlaid for visual inspection. The .csv file will contain well level data (each well occupies a row) summarizing the fraction of cells in each phase of the cell cycle as well as the live and dead cell counts needed for downstream GR calculations. An additional .pdf file can be output at this stage that summarizes the cell cycle data in stacked bar plots per drug (agent).
Usage
The master function run_cell_cycle_gating.run contains a number of arguments and parameters that can be adjusted.
run(data, ndict, dfm=None,
ph3_channel=True, ldr_channel=True,
px_edu=None, x_ldr=None, control_based_gating=False,
control_gates=None, fudge_gates=np.array([0, 0, 0, 0]),
header=7, ldr_gates=None,
nuclei_population_name="Nuclei Selected")
Required arguments
| Name | Description |
|---|---|
data |
The name or path to the input object level data. |
ndict |
A python dict used to map column names in the input data into a format compatible with the script |
Optional arguments
Additional details for some arguments are shown below the table.
| Name | Default | Description |
|---|---|---|
dfm |
None | A dataframe that should contain metadata mapping plates (barcode) and wells to specific experimental conditions such as cell line, drug, and concentration. |
control_based_gating |
False | If True, the script will run automated gating only on those wells that are DMSO controls. Note, a metadata table has to be provided for the script to know which wells are control wells. |
control_gates |
A dataframe that contains control-based (i.e prior) estimates of gating values. | |
fudge_gates |
This argument enables you to offset the automated DNA gates by the provided values. | |
ph3_channel |
True | Set to False if the 4th color channel is not used |
ldr_channel |
True | Set to False if LDR is not used in the assay |
dfm
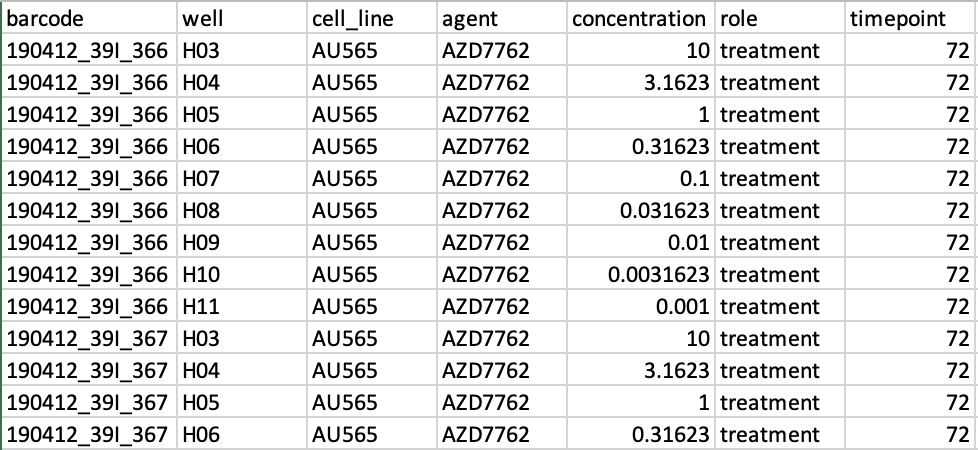
This dataframe can be used to map specific experimental plates to specific experimental conditions. To do this, the dfm input should contain these elements formatted this way.
For example:
fudge_gates
The four inputs to fudge_gates correspond respectively to:
1) the outer left gate 2) the inner left gate 3) the inner right gate 4) the outer right gate
For example: setting fudge_gates to np.array([0, 0.1, -0.2, 0]) will offset the inner gates by 0.1 to the right an 0.2 to the left.

