Overview
Table of contents
What’s ASHLAR?
ASHLAR: Alignment by Simultaneous Harmonization of Layer / Adjacency Registration is a software package that combines multi-tile microscopy images into a high-dimensional mosaic image.
Why ASHLAR?
Recent imaging advances have enabled scientists to cyclically image pathology samples for 20-60 unique proteins, resulting in a tremendous amount of data per pathology slide. These sample areas are generally quite large, so high-magnification images must ‘tile’ to cover the whole slide. Together, this results in highly multiplexed, biologically-rich, image sets that encompass many sample positions and many proteins. To visualize and analyze this data, the images must be combined to seamlessly align adjacent images within cycles and corresponding locations between cycles. To date, this has been a challenge.
ASHLAR is a new software package that fills this need and enables multiplexed image alignment with sub-pixel resolution accuracy.
How does it work?
ASHLAR combines multiplex images through a 3-step process that:
- Stitches together adjacent images within the first imaging cycle,
- registers the relative location of images from subsequent cycles to the first cycle, and
- uses these relative positions to generate a final multidimensional mosaic image.
ASHLAR reads image formats from most commercial microscopes and outputs standard OME-TIFF images. ASHLAR is written in Python and is available under the MIT License. Access it at: https://github.com/labsyspharm/ashlar.
Step 0: Collect multidimensional image data
Collect overlapping image tiles from sample.

Each image tile is assigned an identifier based on sample location. Location data can be extracted directly from BioFormats image metadata (Li et al., 2016) that is produced by many commercial microscopes. ASHLAR can also process images from microscopes that do not support BioFormats if they follow consistent naming convention, acquisition order, and tile overlap.
Note: There are a number of methods to collect multidimensional images. ASHLAR is compatible with many of these methods. For more information on CyCIF, the primary multiplexed image acquisition method used by the Laboratory of Systems Pharmacology at HMS, read the CyCIF manuscript or protocol.
Step 1: Stitching
Align adjacent images across a single cycle by using overlapping regions.
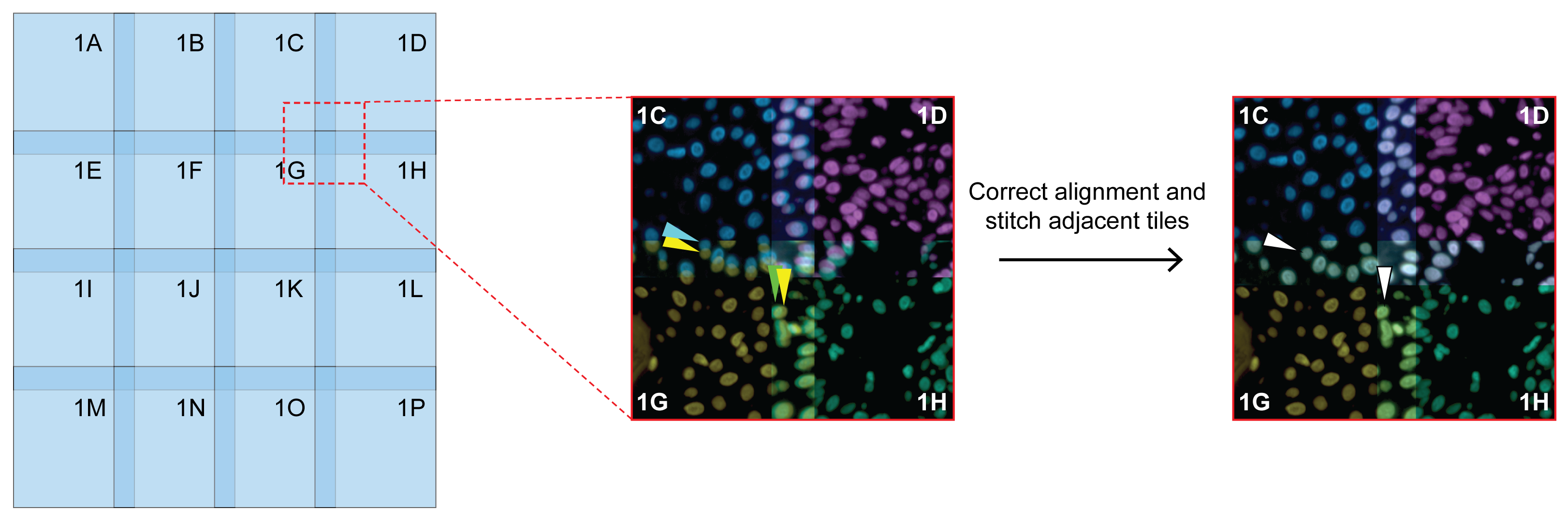
Use one representative channel and propagate locations to other channels per tile. (DNA marker is recommended.)
Step 2: Registration
Align corresponding tiles from each subsequent cycle.
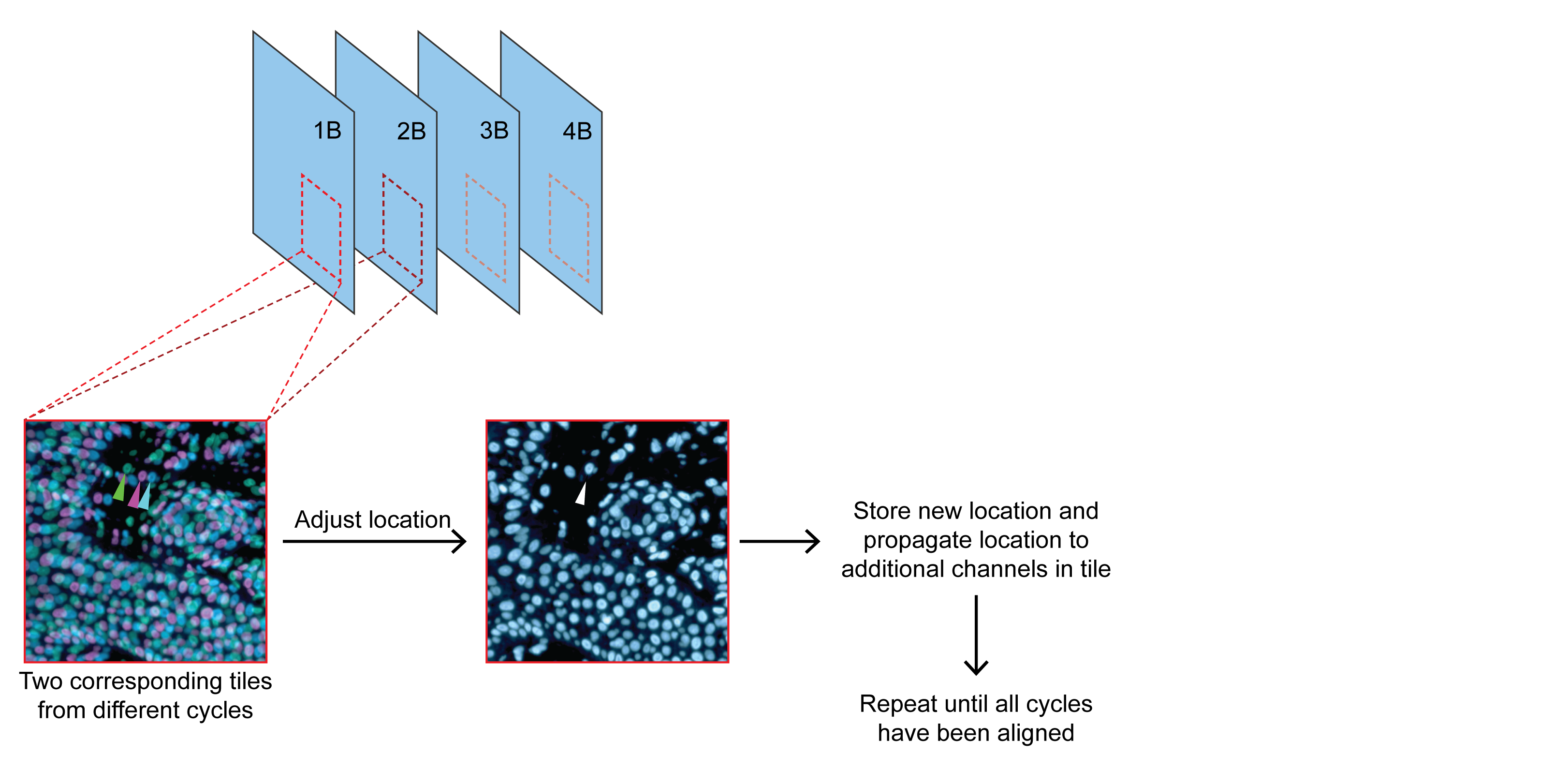
Use one representative channel and propagate locations to other channels per tile. (DNA marker is recommended.)
Step 3: Mosaic Image Generation
All corrected tile positions from Step 1 (stitching) and Step 2 (registration) are combined.

Learn More
View the detailed computational methods for more information on how each step is performed.
For more details, read the preprint manuscript here: https://doi.org/10.1101/2021.04.20.440625.
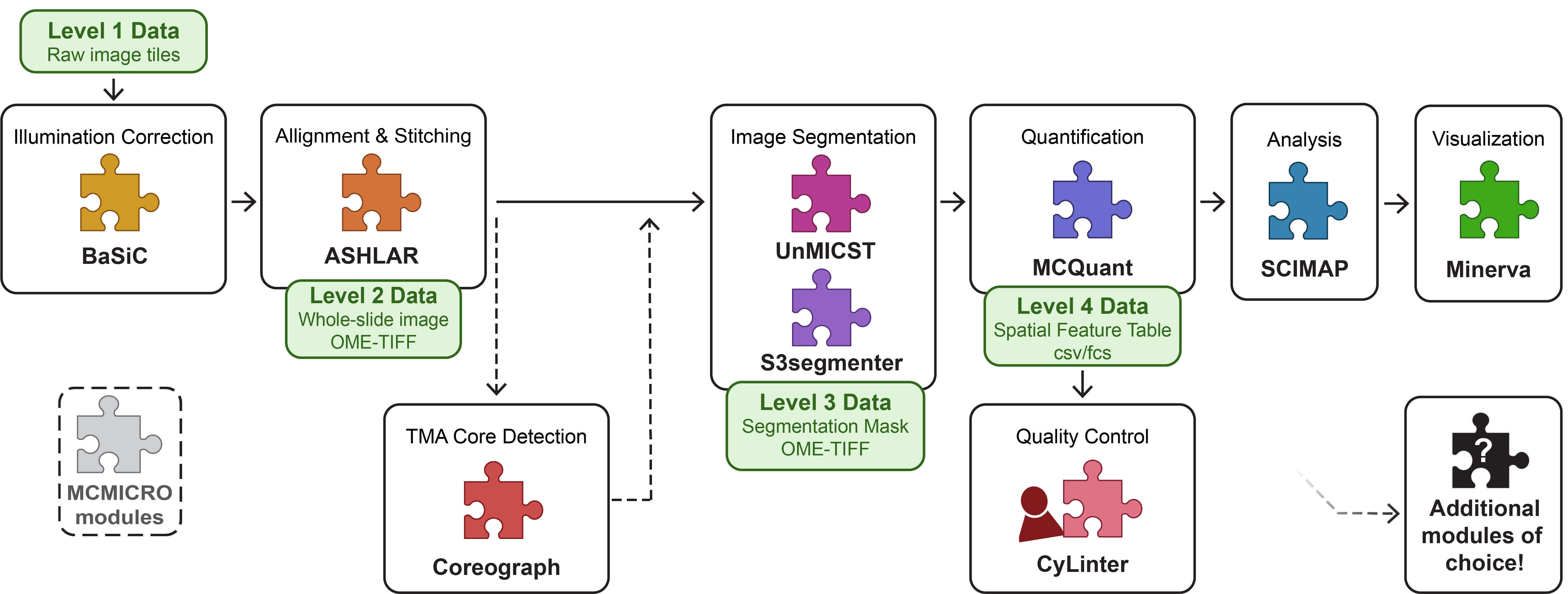
Integration with MCMICRO Pipeline
The Multiple-choice microscopy pipeline (MCMICRO) is an end-to-end processing pipeline to transform large, multi-channel whole slide images into single-cell data. See https://mcmicro.org/ for more information on the MCMICRO pipeline documentation, implementation, troubleshooting, and more.