This website accompanies the manuscript:
Integrating multi-omics data
reveals function and therapeutic potential
of deubiquitinating enzymes
(2021) Laura M Doherty, Caitlin E Mills, Sarah A Boswell, Xiaoxi Liu, Charles Tapley Hoyt, Benjamin M
Gyori, Sara J Buhrlage, and Peter Karl Sorger.
eLife, 11:e72879.
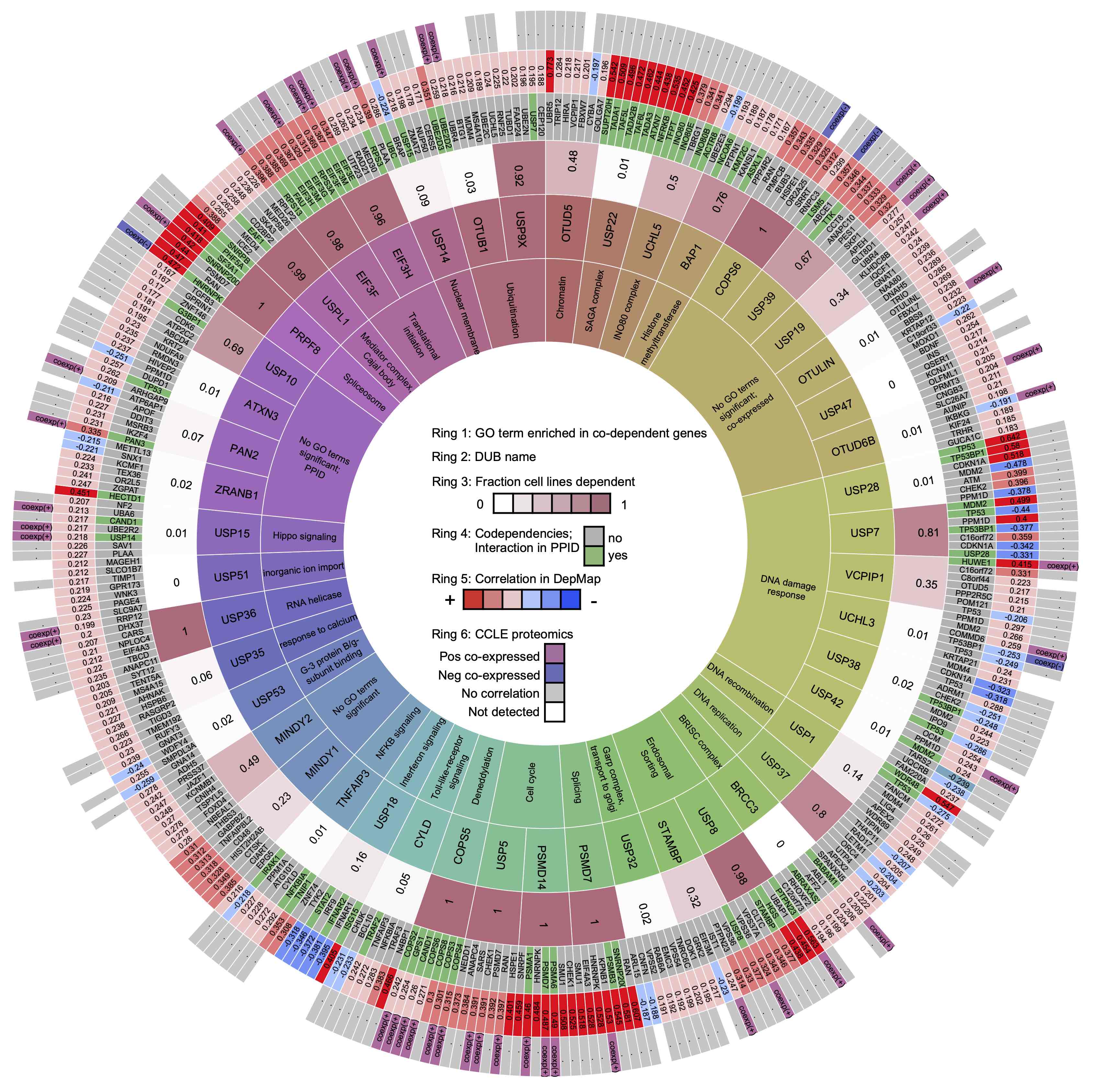
Doherty et al. analyze deubiquitinating enzymes (DUBs) for relationships with other genes in the Cancer Dependency Map, Connectivity Map, Cancer Cell Line Encyclopedia, and protein-protein interaction databases: BioGRID, Pathway Commons, Reactome, IntAct, and NURSA. The DUB Portal presents these results in an interactive form, extends them with additional literature context, and links them directly to outside resources.
Landing page
The landing page at https://labsyspharm.github.io/dubportal/ shows a table of 99 DUBs analyzed in Doherty et al. with some key properties and metrics associated with them. The column of the table can be interpreted as follows:
- DUB
- The official HUGO Gene Nomenclature Consortium (HGNC) gene symbol of the DUB gene
- DUB name
- The gene name associated with the DUB as provided by HGNC
- DUB family
- The family to which a given DUB belongs in the FamPlex ontology
- # Papers
- The number of publications about the DUB indexed in PubMed, as determined by a search using the PubMed API. The number in this column also links to PubMed where the set of papers can be browsed. (Note: the count displayed on the DUB Portal and the PubMed website can differ depending on the actual state of the PubMed web service)
- # DUB Stmts
- The number of distinct statements assembled by INDRA (indra.bio) by text mining the literature and processing pathway databases in which the given DUB is reported to deubiquitinate a target.
- # Other Stmts
- The number number of distinct statements assembled by INDRA (indra.bio) by text mining the literature and processing pathway databases in which the given DUB appears in any role, except for statements counted under # DUB Stmts.
- Dependent Cell Lines
- The percentage of cell lines that show dependency for the given DUB per the Cancer Dependency Map.
- # Sig. ORA Gene Sets (DepMap)
- The number of gene sets considered significant (p < 0.05) by over-representation analysis with multiple hypothesis testing correction using genes depending on a given DUB as the query set.
- DGEA
- How many genes were significantly differentially expressed due to knockout of the given DUB
- # Sig. GSEA Gene Sets (DGEA)
- The number of gene sets considered significant (p < 0.05) by GSEA analysis with multiple hypothesis testing correction using genes differentially expressed by knocking out the given DUB as the query set
By clicking on the DUB’s gene symbol or name, a page is displayed with detailed information specific to the given DUB.
DUB-specific pages
In the header, the DUB’s official gene symbol and name are listed along with links to a landing page for the given DUB and its orthologs across outside resources including HGNC, Entrez Gene, UniProt, MGI, and RGD. Links to Pathway Commons as well as the INDRA Database are also shown allowing browsing interactions associated with the DUB.
The DepMap Analysis section summarizes the method used in the analysis in Doherty et al. and then shows the table of seven top-correlated genes for the given DUB. The table provides the DepMap Correlation for each gene and Evidence from multiple sources that provide support for the given correlation in the form of prior knowledge about protein interactions. The evidence column contains badges for each interaction source (e.g., BioGRID, INDRA), in some cases also linking to a relevant page in the given resource.
The Dependency GO Term Enrichment section shows GSEA analysis results on the genes correlated using terms from the Gene Ontology and gene sets derived from the Gene Ontology Annotations database via MSigDB. Results below a p-value of 0.05 are shown in a table. The table contains the following columns:
- GO identifier
- The identifier for the enriched GO term
- GO Name
- The standard name for the enriched GO term
- GO Type
- The type of GO term (Biological Process, Cellular Component or Molecular Function)
- p-value, adjusted p-value, and q-value
- The raw and adjusted p-value and the q-value associated with the enrichment.
The Literature Mining section extends on Doherty et al. to provide additional context on a given DUB, assembled from the scientific literature and pathway databases using the INDRA knowledge assembly system. Statements specific to the given DUB's deubiquitinase activity are shown in the first list, and all other Statements involving the DUB are shown in the second list. Both statement lists consist of a list of headings with each heading representing an interaction or regulation relationships such as “ATXN3 deubiquitinates STUB1”. Clicking on each heading opens up a list of specific evidence sentences from the literature supporting that statement with the names of entities highlighted and with links to the source publication. Statements can also be curated for correctness. To do this, click on the arrow to the left of the name of the source of the evidence to link out to the INDRA Database website where curations can be submitted as described in this tutorial. Statements curated as incorrect are excluded when the DUB Portal is rebuilt.
Maintenance
The DUB Portal is built and kept up to date automatically on a weekly basis using Github Actions workflows running Python code available at https://github.com/labsyspharm/dubportal. The number of publications as well as the number and list of INDRA Statements for each DUB are updated with each build to pull the latest available content.
Versioning
- HGNC*
- 2023-02-19
- INDRA*
- 2023-02-19
- PubMed*
- 2023-02-19
- NURSA
- static
- BioGRID*
- 4.4.218
- DepMap*
- 22Q4
- Pathway Commons*
- Reactome*
- 83
Resources with an asterisk are automatically updated.
Contact
The DUB Portal was built by the INDRA team (indralab.github.io), part of the Laboratory of Systems Pharmacology, Harvard Medical School. Please contact indra.sysbio@gmail.com or submit an issue at https://github.com/labsyspharm/dubportal with questions or comments.