This DUB portal (which updates automatically each week) provides the integration and analysis of publicly available resources as well as newly collected transcriptomic data following deubiquitinating enzyme (DUB) knockout or inhibition to facilitate the exploration of DUB function in oncology.
This website accompanies the manuscript: Integrating multi-omics data reveals function and therapeutic potential of deubiquitinating enzymes
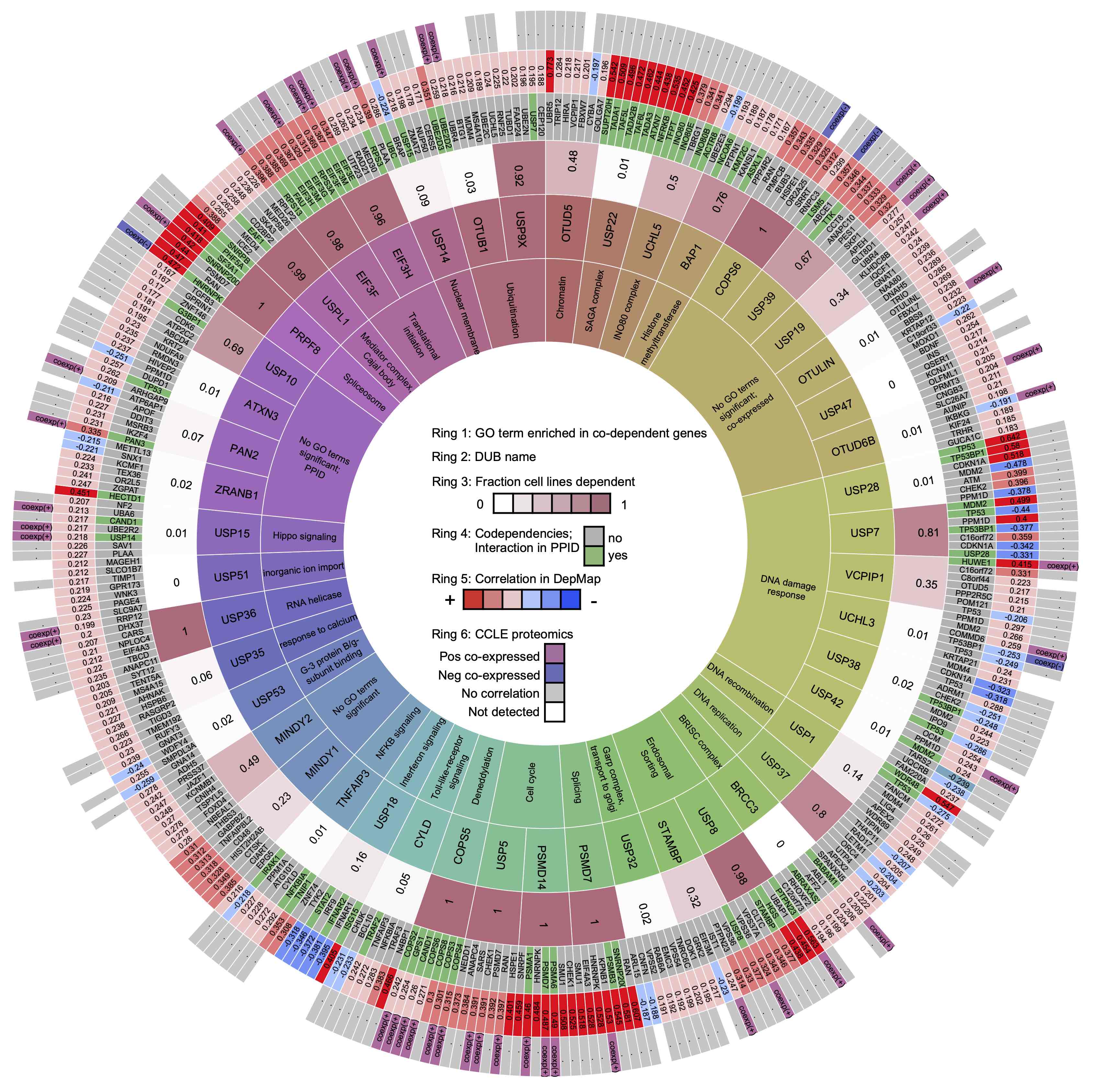
To assemble a knowledgebase of DUB activities, co-dependent genes, and substrates, we combined targeted experiments using CRISPR libraries and inhibitors with systematic mining of functional genomic databases. Analysis of the Dependency Map, Connectivity Map, Cancer Cell Line Encyclopedia, and protein-protein interaction databases yielded specific hypotheses about DUB function, a subset of which were confirmed in follow-on experiments.
The data in this study, which are browsable below, promise to improve understanding of DUBs as a family as well as the activities of specific DUBs, which have been targeted with investigational cancer therapeutics.
Deubiquitinating enzymes (DUBs) are proteases that remove ubiqutin conjugates from proteins, thereby regulating protein turnover. However, the majority of substrates and pathways regulated by DUBs remain unknown, impeding efforts to prioritize specific proteins for research and drug-development.
Each DUB is listed with the number of papers in PubMed, the number of INDRA statements about deubiquitinating substrates, the number of INDRA statements total (including upstream and downstream processes beyond deubiquitination), and the and the fraction of cell lines in the DepMap that are strongly dependent on each DUB (i.e., cancer cell proliferation is strongly impacted by DUB knockout). Blank values in the "Dependent Cell Lines" column means the DUB was not knocked out in the DepMap dataset. Additionally, we list the Gene Ontology (GO) term most strongly enriched in the top seven co-dependent genes for each DUB and the corresponding p-values. Finally, the "DGEA" column contains the number of differentially expressed genes 96 hours following knockout of the particular DUB in MDAMB231 cells as measured by high-throughput RNAseq. Blank values in "DGEA" refer to DUBs that were not knockout out in the RNAseq experiment.
Click on an individual DUB name for more details and additional data and analysis.