Insight into the DUB family as a whole
The majority of DUBs have non-redundant functions
It has been suggested that redundancy among DUBs (Vlasschaert et al., 2017) might limit the effectiveness of selective DUB inhibitors as therapeutic agents, (Davis and Simeonov, 2015). However, we find that single gene knockouts of 43 DUBs impact proliferation in at least 8 DepMap cancer cell lines, and 21 DUBs are embryonic lethal with complete or partial penetrance in mice; deletion of an additional 26 DUBs has a scorable murine phenotype. Thus, many DUBs appear to have non-redundant functions. Moreover, since many targets for successful anticancer drugs are embryonic lethal, (Yu and Xu, 2020) our data support further development of DUBs as cancer therapeutics.
DUBs as E3 Regulators
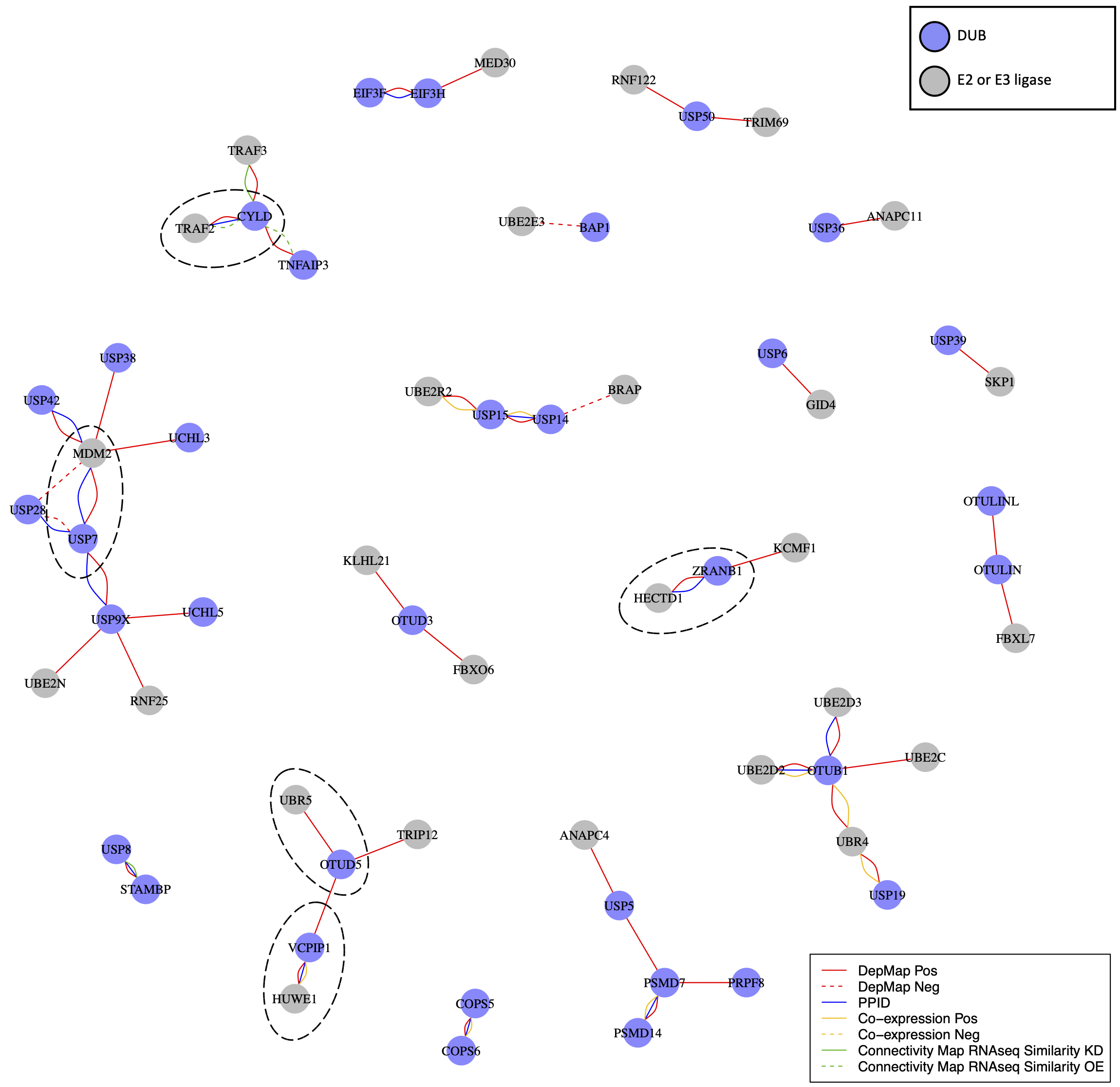
USP7 is a regulator of MDM2, the E3 ligase for the TP53 tumor suppressor protein. The relationship between USP7 and MDM2 does not appear to be the only instance of a DUB regulating an E3 ligase. DepMap co-dependent genes for DUBs were strongly enriched and positively correlated with E3 ligases and other ubiquitin or ubiquitin-like transferases; in many cases, DepMap data were supported by PPID or co-expression data (e.g. the VCPIP1 DUB and HUWE1 E3 ligase; the DUB ZRANB1 and the HECTD1 E3 ligase).
DUBs are expected to antagonize E3 ligase activity by deubiquitinating E3 ligase substrates, making negative correlations the expected outcome. However, it has also been suggested that DUBs might associate directly with E3 ligases and inhibit their auto-ubiquitination activity, thus preventing proteasomal degradation of the E3 ligase (Wilkinson, 2009). In this case, positive correlations in the DepMap between E3 ligases and DUBs would be expected. We found that multiple DUBs in fact exhibited strong positive rather than negative correlations with one or more E3 ligases in the DepMap data.
Overall, we identified 23 DUBs with at least one co-dependent E3 ligase, and 8 of these DUBs had a co-dependent E3 ligase also supported by PPID or co-expression data. Selected DUBs have previously been reported to stabilize E3 ligases; for example, USP7 stabilizes MDM2, CYLD stabilizes TRAF2, and OTUD5 stabilizes UBR5 (de Vivo et al., 2019; Lork et al., 2017). However, our data suggest that this may be a general feature of the DUB family, with many E3 ligases interacting with DUBs that antagonize E3 auto-ubiquitination and increase protein stability (Wilkinson, 2009). Multiple E3 ligases act as oncogenes, and promoting their degradation via DUB inhibition may be a broadly useful therapeutic strategy.
DUB E3 ligase network: Ubiquitin or ubiquitin-like transferases whose co-dependency relationships correlated with DUBs in the DepMap. DUBs are colored blue and ubiquitin transferases are colored grey. Red lines represent correlations in the top seven co-dependent genes. Green lines represent similarity by CMap (tau similarity score > 90). Yellow lines represent co-expression in proteomics (FDR < 0.01 and |z-score| > 2). Blue lines represent interaction in protein-protein interaction databases.
Comparing DUB inhibitors and DUB knockouts
There is growing interest in developing small molecule DUB inhibitors for use as human therapeutics (Davis and Simeonov, 2015; Harrigan et al., 2018) but the field is still relatively new. Early generation DUB inhibitors were not included since many of these have been shown to have substantial polypharmacology (Altmann et al., 2017; Kluge et al., 2018; Schauer et al., 2020; Schlierf et al., 2016).
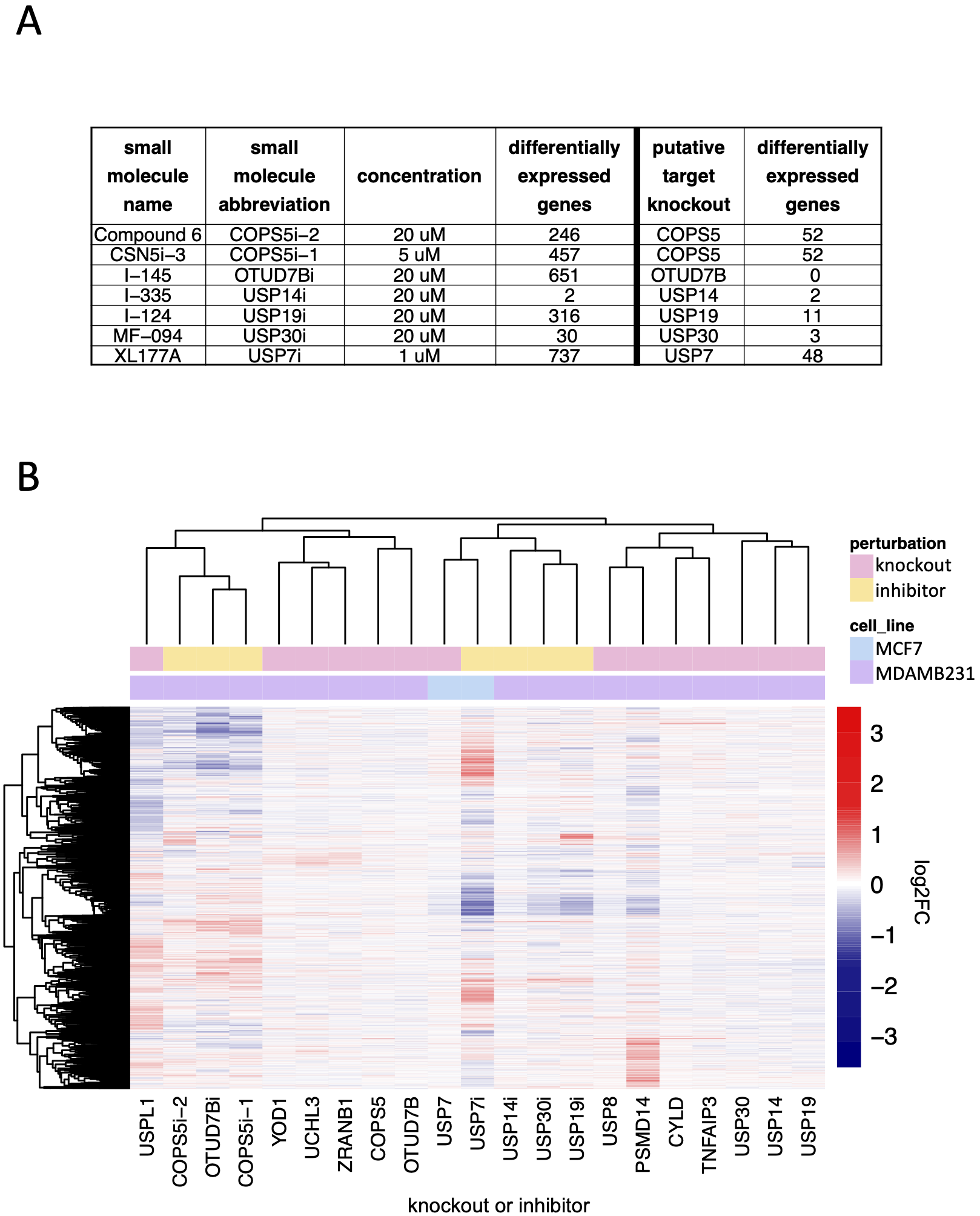
Using transcript profiling we compared seven small molecules reported by their developers to be highly selective inhibitors of specific DUBs to CRISPR-Cas9 mediated knockout of their targets. DUB inhibitor signatures were significantly similar to knockout signatures in only two cases: USP14 inhibition with I-335 and USP7 inhibition with XL177A. We conclude that these compounds are selective, although the signature of USP7 inhibition was substantially stronger than that of USP7 knockout. This was true in general, with exposure of cells to DUB inhibitors resulting, in all cases, in significantly more DE genes than knockouts. In the case of XL177A, our studies cannot determine whether this difference reflects the time at which the measurements were made, the degree of USP7 inhibition by drug or mRNA depletion by CRSPR-Cas9, or the existence of off-target effects. In the cases of the COPS5, OTUD7B, and USP30 inhibitors (inhibition of COPS5 with Compound 6 or CSN5i-3, inhibition of OTUD7B with I-145, inhibition of USP19 with I-124, and inhibition of USP30 with MF-094), the lack of significant correlation between the generally weak knockout phenotypes and the strong drug-induced phenotypes suggest substantial off-target activity. Small molecules targeting multi-protein families via competitive inhibition at the active site commonly exhibit some degree of polypharmacology (that is, they exert their biological effects by binding to multiple targets)(Giri et al., 2019). It appears that, except in the case of USP7 and USP14, additional medicinal chemistry will be required to manage polypharmacology.
Comparison of DUB knockout and inhibition: (A) The number of significantly differentially expressed genes (adjusted p-value < 0.05) as a result of small molecule DUB inhibition (24 hours treatment) and knockout of the putative target (96 hours after transfection with guide). (B) Hierarchical clustering of log2FC values for significantly differentially expressed genes (adjusted p-value < 0.05) for small molecule inhibitors of DUBs, the knockout of the putative DUB targets of the small molecules, and the DUB knockout hits that resulted in more than 20 differentially expressed genes.