Discriminating among essential and non-essential DUB functions for well-studied DUBs
In several cases, our studies yielded unexpected hypotheses about the functions of DUBs that have already been well studied.
USP14
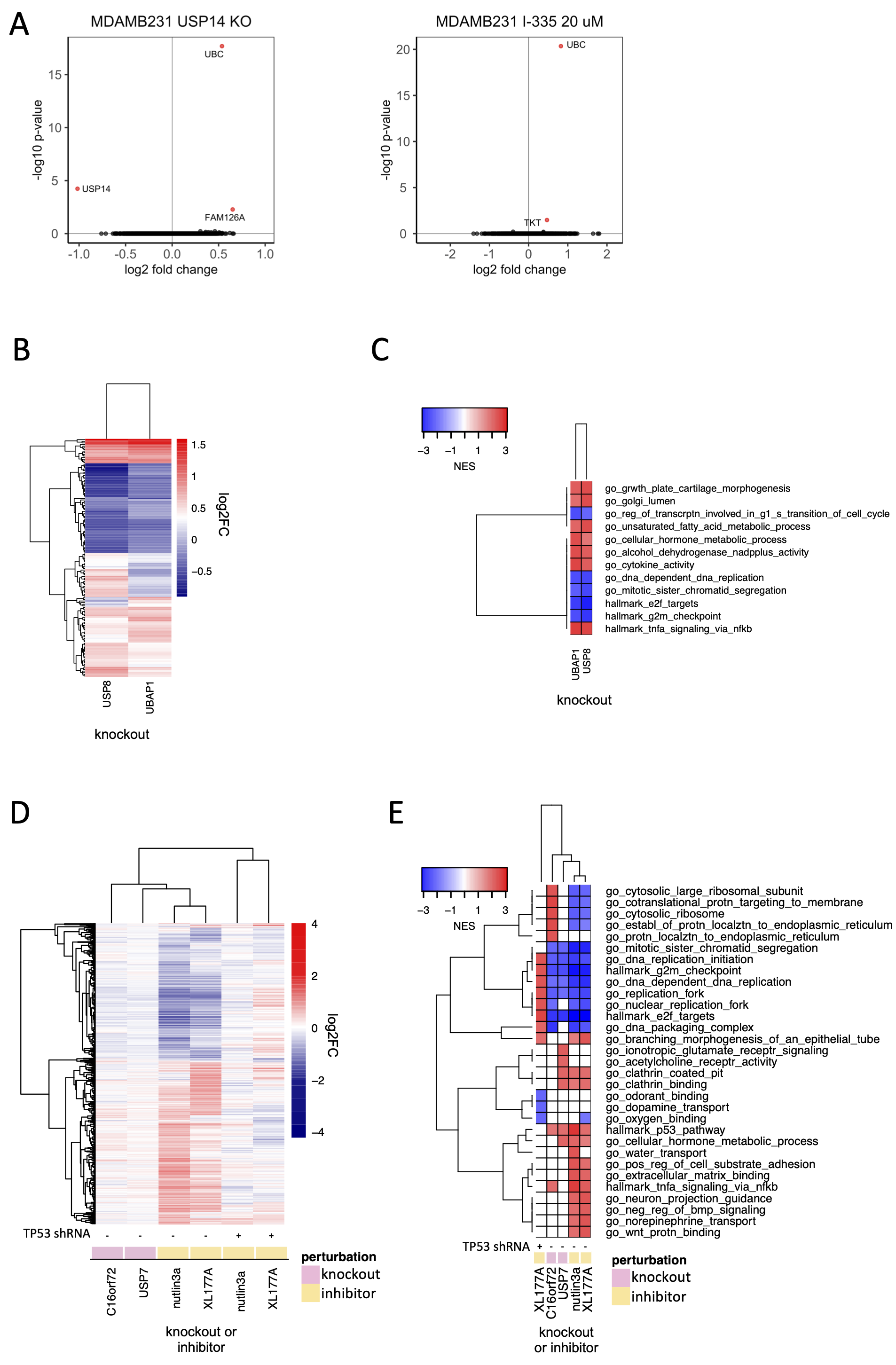
USP14 is a component of the proteasome; however, we found that USP14 was strongly co-dependent in DepMap data not with subunits of the proteasome but instead with the UBC polyubiquitin gene, a primary source of ubiquitin in mammalian cells. Knockout of USP14 by CRISPR-Cas9, or exposure of cells to the USP14 inhibitor I-335 resulted in highly selective upregulation of the UBC gene. We therefore propose that maintenance of the pool of free ubiquitin, not regulation of the proteasome, is likely to be the key, non-redundant function for USP14.
UCHL5
UCHL5 is a component of both the INO80 complex and the proteasome. The effect of UCHL5 knockout is most similar to that of knockout of other INO80 subunits (e.g. NFRKB, TFPT, INO80, INO80E, INO80B, r range = 0.29 to 0.54) and no significant correlation was observed in DepMap data with knockout of proteosome subunits (e.g. PSMD9, PSMD6, PSMD3, r range = 0.03 to 0.05). We hypothesize that UCHL5 plays an essential and non-redundant function in the INO80 complex rather than the proteasome. Since the INO80 complex has an essential role in DNA damage repair (Yao et al., 2008), UCHL5 inhibitors may be most useful in combination with DNA damaging agents.
USP8
USP8 is an extensively studied DUB that has been shown to stabilize endosomal sorting complexes required for transport (ESCRT) and receptors. We find that knockout of USP8 also upregulates the expression of cytokines, implying that USP8 may have a role in recycling cytokine receptors as well as growth factor receptors. We knocked out three ESCRT proteins (UBAP1, HGS, PTPN23) that strongly correlated with USP8 in DepMap data. We found that the transcriptional signature of UBAP1 knockout was strongly correlated with that of USP8 knockout and that both knockouts resulted in upregulation of multiple cytokines. In DepMap data, USP8 correlated more strongly with ESCRT machinery proteins than with individual growth factor or cytokine receptors, independent of cancer lineage, suggesting that the essential function of USP8 in cancer cells is not mediated by one specific receptor alone – e.g. EGFR – but rather by multiple growth factor and cytokine receptors that undergo similar ESCRT-dependent endosomal sorting.
USP7
One of the most promising potential uses of DUB inhibitors is to indirectly regulate the levels of disease-associated genes that are not conventionally considered to be druggable such as transcription factors and scaffolding proteins. This strategy has been most actively pursued for USP7, which is a regulator of MDM2, the E3 ligase for the TP53 tumor suppressor protein: inhibition of USP7 increases the levels of ubiquitinated MDM2, promoting its degradation and thereby increasing TP53 levels.
Our data on USP7 are consistent with this hypothesis: we find that the top DepMap co-dependent gene for USP7 is MDM2, and knockdown of TP53 largely rescues the transcriptional phenotype observed for USP7 inhibition in TP53 wildtype cells. We find that USP7 has at least one additional substrate, C16orf72, that may also be a TP53 regulator. Moreover, the USP7 inhibitor XL177A has a phenotype that is independent of TP53 and involves upregulation of histone genes and genes involved in the G2M cell cycle checkpoint. We speculate that this may reflect the reported involvement of USP7 in the regulation of polycomb complexes (de Bie et al., 2010), although we cannot rule out an off-target activity for XL177A. We conclude that the primary role of USP7 in cancer cells involves the MDM2-TP53 axis (Schauer et al., 2020).
Discriminating the functions of well-studied DUBs: (A) Changes in gene expression (log2 fold change vs. log 10 adjusted p-value, adjusted p-value < 0.05 colored red) in MDAMB231 cells 96 hours after USP14 knockout by CRISPR-Cas9 (left) and 24 hours after treatment with the USP14 inhibitor I-335 at 20 µM (right). (B) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value < 0.05) 96 hours following knockout of USP8 or UBAP1 in MDAMB231 cells. (C) Gene sets significantly (FDR < 0.05) enriched in MDAMB231 cells 96 hours after knockout of USP8 or UBAP1. The top five upregulated and top five downregulated gene sets for each condition are shown. (D) Hierarchical clustering of significantly differentially expressed genes (adjusted p-value < 0.05) 96 hours after knockout of USP7 or C16orf72 in wild-type MCF7 cells or following a 24-hour treatment with 5 µM nutlin3a or 1 µM XL-177A in wildtype or p53 knockdown MCF7 cells. (E) Gene sets significantly enriched (FDR < 0.05) for the conditions shown in (D). The top five up- and down-regulated gene sets for each condition are shown.