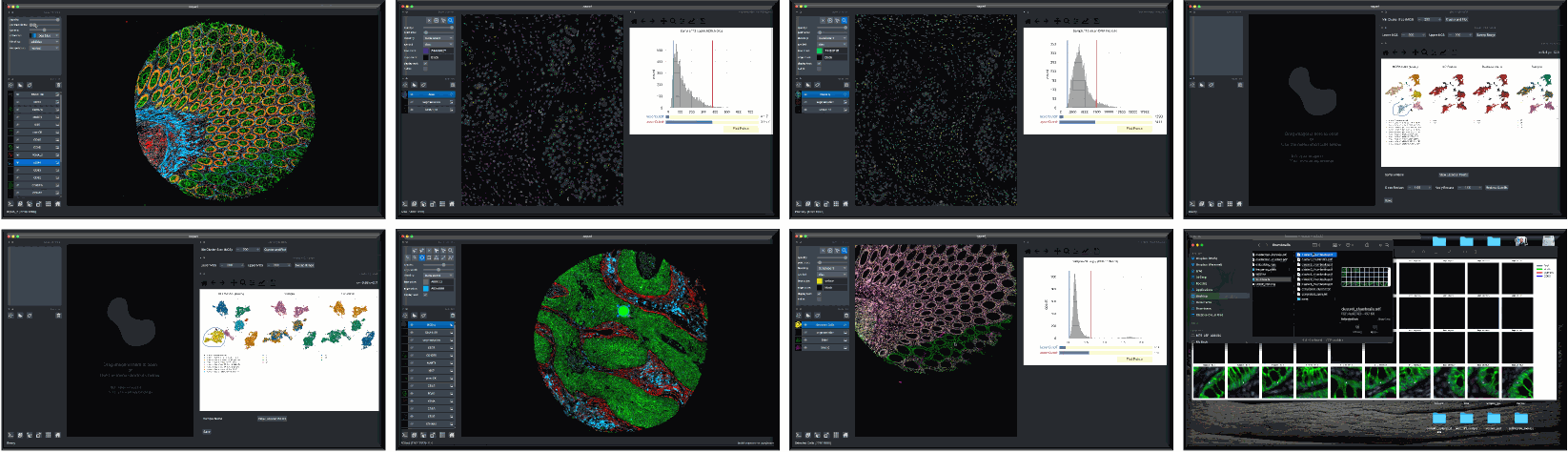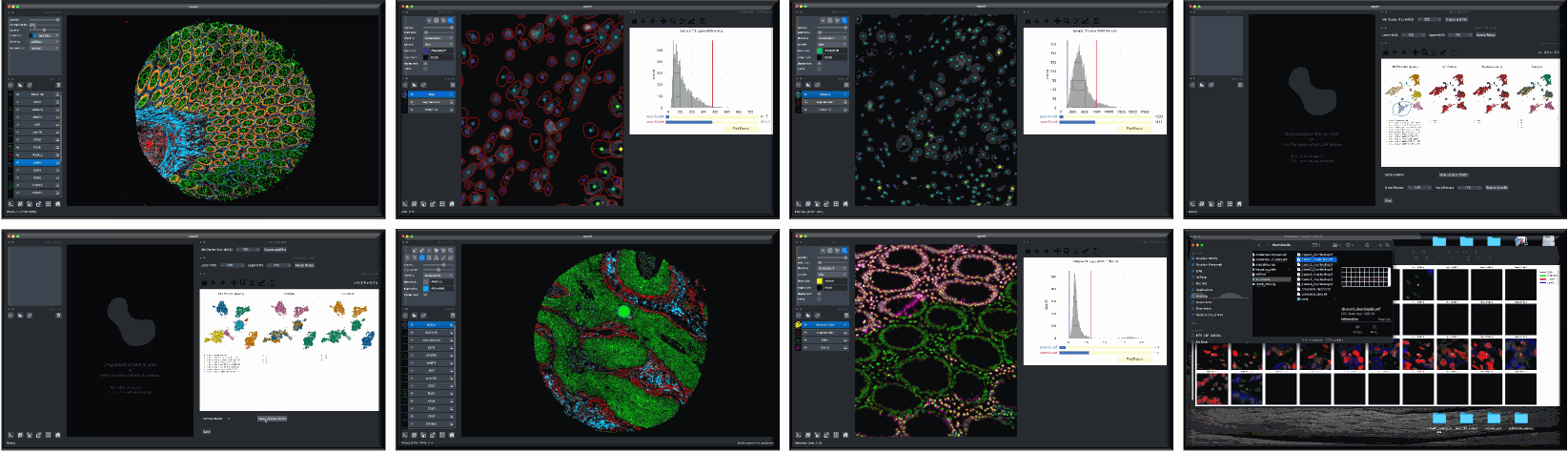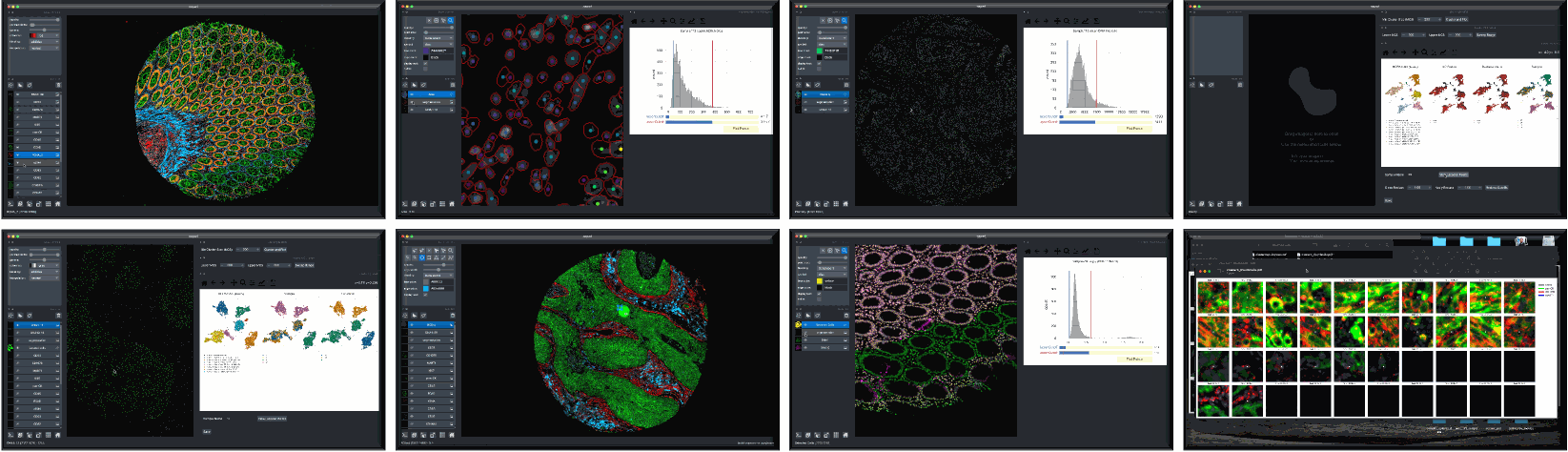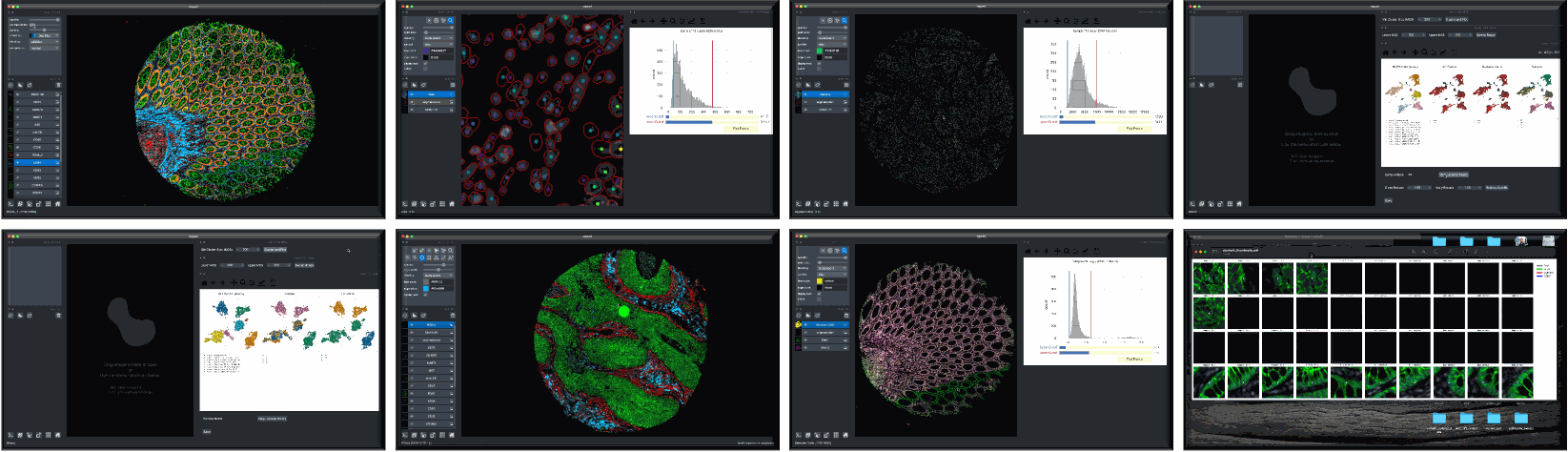
Overview
QC for Multiplex Microscopy
Although quality control (QC) methods have long been associated with analysis tools for single-cell genomics and transcriptomics research, analogous tools have lagged in the area of quantitative microscopy. There are now at least 9 different multiplex imaging platforms capable of routine acquisition of 20-40 channel microscopy data1,2,3,4,5,6,7,8,9 and each is sensitive to microscopy artifacts. Current tools for microscopy-based QC act on pixel-level data10,11,12,13,14. CyLinter differs in that it allows users to work with both pixel-level and single-cell data to identify and remove cell segmentation instances in multiplex images corrupted by visual and image-processing artifacts that can dramatically affect data interpretation.
About CyLinter
CyLinter is open-source QC software for multiplex microscopy. The tool is instantiated as a Python Class and consists of multiple QC modules through which single-cell data are passed for serial redaction. Partially-redacted spatial feature tables are cached within and between modules to allow for iterative QC strategies and progress bookmarking. CyLinter is agnostic to data acquisition platform (CyCIF1, CODEX2, MIBI3, mIHC4, mxIF5, IMC6, etc.) and takes standard TIFF/OME-TIFF imaging files and CSV spatial feature tables as input.
- Lin, J.-R. et al. Highly multiplexed immunofluorescence imaging of human tissues and tumors using t-CyCIF and conventional optical microscopes. Elife 7, (2018).
- Goltsev, Y. et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968-981.e15 (2018).
- Angelo, M. et al. Multiplexed ion beam imaging (MIBI) of human breast tumors. Nat Med 20, 436–442 (2014).
- Tsujikawa, T. et al. Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Reports 19, 203–217 (2017).
- Gerdes, M. J. et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci U S A 110, 11982–11987 (2013).
- Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 11, 417–422 (2014).
- Remark, R. et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Science Immunology 1, aaf6925–aaf6925 (2016).
- Gut, G., Herrmann, M. D. & Pelkmans, L. Multiplexed protein maps link subcellular organization to cellular states. Science 361, (2018).
- Saka, S. K. et al. Immuno-SABER enables highly multiplexed and amplified protein imaging in tissues. Nat Biotechnol 37, 1080–1090 (2019).
- Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M. & Madabhushi, A. HistoQC: An Open-Source Quality Control Tool for Digital Pathology Slides. JCO Clin Cancer Inform 3, 1–7 (2019).
- Ameisen, D. et al. Towards better digital pathology workflows: programming libraries for high-speed sharpness assessment of Whole Slide Images. Diagn Pathol 9 Suppl 1, S3 (2014).
- Senaras, C., Niazi, M. K. K., Lozanski, G. & Gurcan, M. N. DeepFocus: Detection of out-of-focus regions in whole slide digital images using deep learning. PLoS One 13, e0205387 (2018).
- Wen, S. et al. A Methodology for Texture Feature-based Quality Assessment in Nucleus Segmentation of Histopathology Image. J Pathol Inform 8, 38 (2017).
- Baranski, A. et al. MAUI (MBI Analysis User Interface)-An image processing pipeline for Multiplexed Mass Based Imaging. PLoS Comput Biol 17, e1008887 (2021).

